
If you open a VCF file that does not contain genotypes data, the view will be different, displaying only the bars marking the calls, as shown below. You can change the height of the rows in the window provided. You can also adjust the height of the squished row by right-clicking and selecting Change Squished Row Height. Squished shows the genotypes with the rows compressed to maximize the data visible on the page. The Display Mode changes what you can see of the data:Ĭollapsed removes all the genotypes, leaving only the allele frequency bars.Įxpanded shows the genotypes at the usual row height, with the sample names in the first column.

You can select the sort twice for the same variant to flip it, i.e., if you sort depth, it sorts from high to low select the depth sort a second time to sort from low to high. The Sort Variant By options allow you to sort the set by a trait of a specific variant. To change the color coding of the plot, select Color By>Allele. Functions are provided to rapidly read from and write to VCF files. Facilitates easy manipulation of variant call format (VCF) data. To change this, right-click and select Set Feature Visibility Window. The R package vcfR is a set of tools designed to read, write, manipulate and analyze VCF data. The window size at which VCF data is loaded is proportional to the number of samples. Find more details on the menu options on the Pop-up Menu page. Some of the options are specific to the variant selected. To see the options for changing the view of your VCF file, right-click on a variant. If a file has more than 10 genotypes, the VCF file will be opened in its own pane, with a scroll bar, as shown below. It is a simple VCF viewer in which you can add multiple VCF files and view all contacts and.
VCF FILE VIEWER SOFTWARE
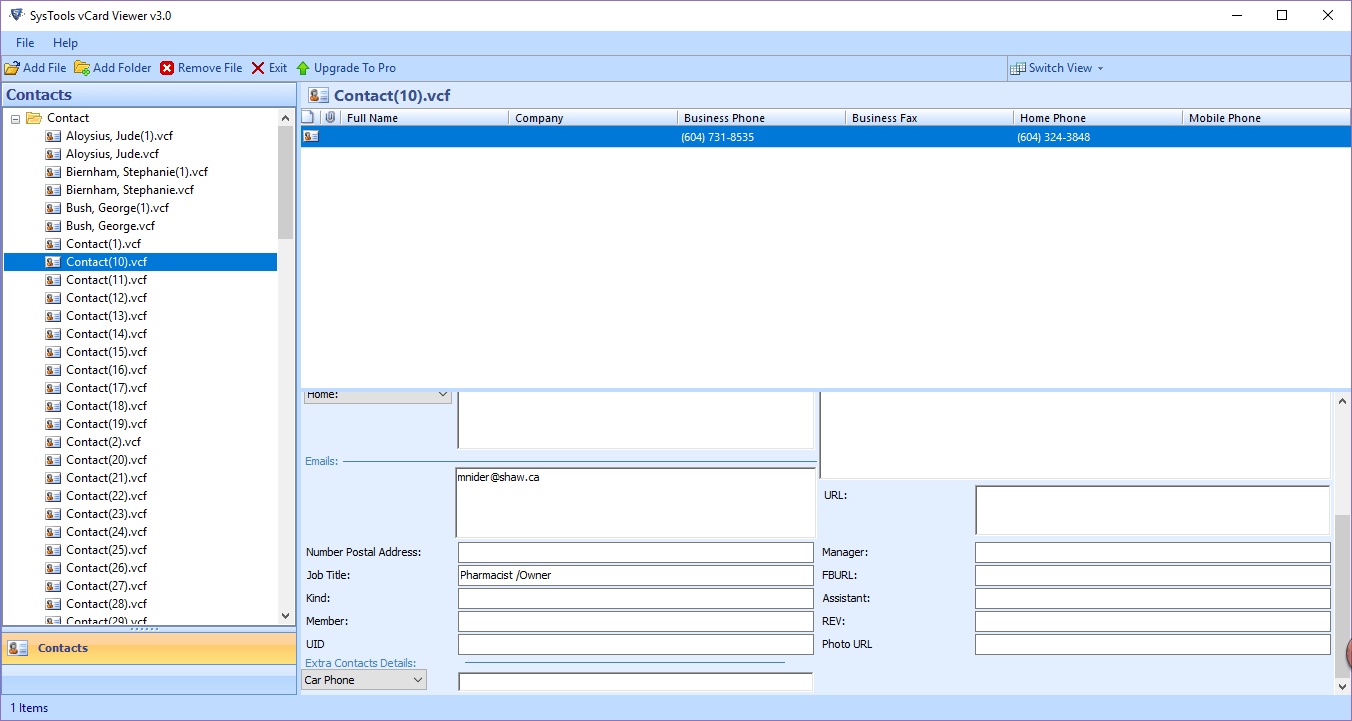
Dark blue = heterozygous, Cyan = homozygous variant, Grey = reference. SysTools vCard Viewer is a free VCF viewer software for Windows. The genotypes for each locus in each sample. The format is further described on the 1000 Genomes project Web site.Įach bar across the top of the plot shows the allele fraction for a single locus. VCF stands for Variant Call Format, and this file format is used by the 1000 Genomes project to encode SNPs and other structural genetic variants.


 0 kommentar(er)
0 kommentar(er)
